本資料は2019年10月31日に社内共有資料として展開していたものをWEBページ向けにリニューアルした内容になります。
■Computational approaches to study Genetics



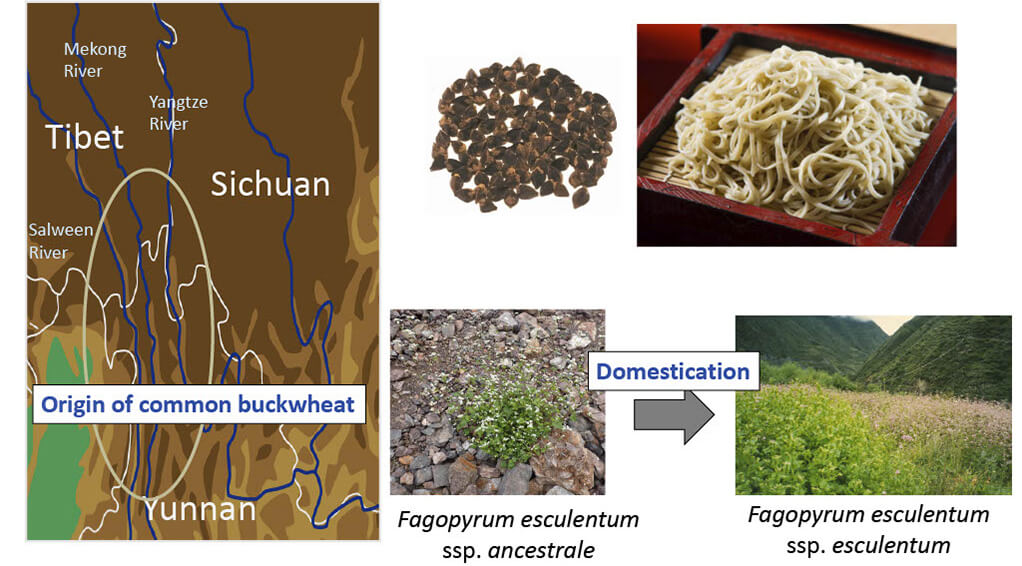

■Aim of Buckwheat Project
- Collect & maintain genetic resources of buckwheat-related species
(many wild species are facing extinction due to development) - Understand domestication process of buckwheat
- Identify genes important for breeding of buckwheat
“Extensive characterization of domestication-related genes in buckwheat by utilizing the genetic resource of Yunnan province, China”
KAKENHI Fostering Joint International Research B, PI: Yasuo Yasui (Kyoto U)
中国雲南省の野⽣ソバ遺伝資源を活⽤した栽培化関連遺伝⼦の網羅的同定 科研費
・ 国際共同研究強化B 研究代表者:安井康夫(京都⼤)
■Why am I involved?

- Huge advance in technology to generate large-scale DNA data
- Need for people with computational skills and knowledge of genetics
(very few such people that work on buckwheat, horses, etc)

■Outline
- Basic concepts of genetics and evolution
- What is written in the DNA?
- How can we know what’s written in the DNA?
- How can we associate “genotype” with “phenotype”?
- Research on Thoroughbred horses
■History of Abstraction in Biology
Biology is… complex, diverse, changes over time
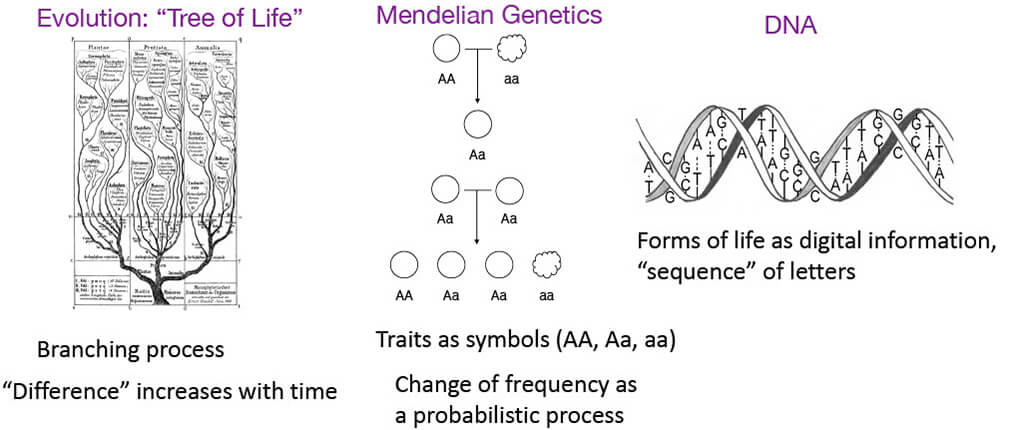
■Basic concepts of genetics
- Parent and offspring are similar – genetic information is passed on What is passed on and how?
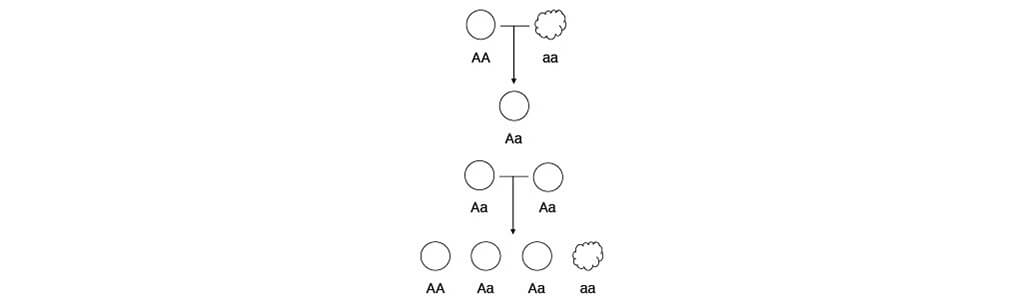
- Information of A and a is transmitted across generations without changing
- The “phenotype” (appearance) of AA and Aa are the same
■Mechanism of Heredity

Genome: entire set of genetic information Every human has 2 genomes (1 from each parent)
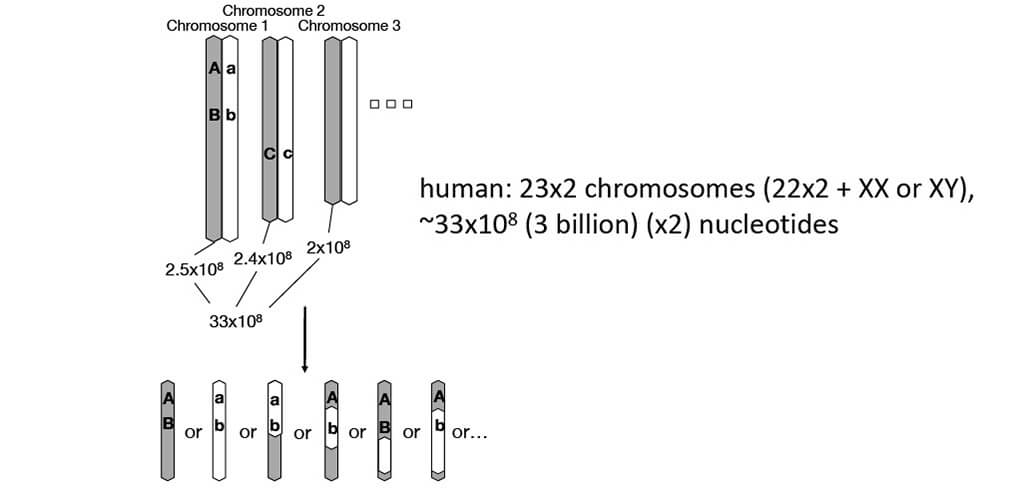
■Large variation in genome size

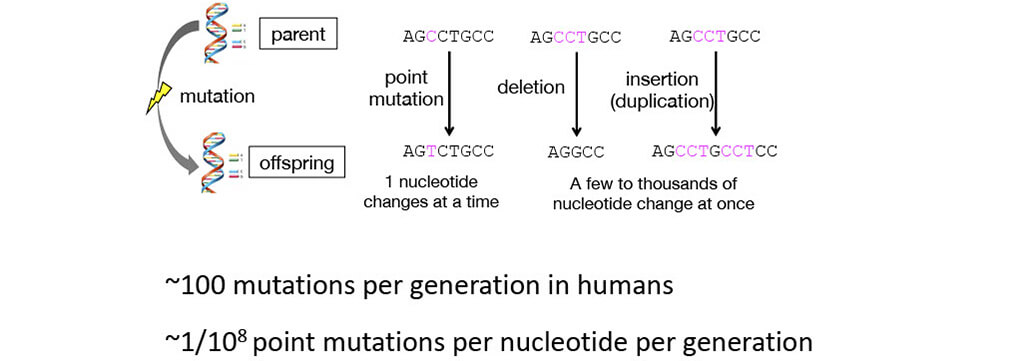
■Mutation
“Replication” cannot generate diversity, genetic information changes
■Evolution
- Heredity
- Mutation => some of them change the “phenotype”
- “Population process” (competition, “struggle for survival”)
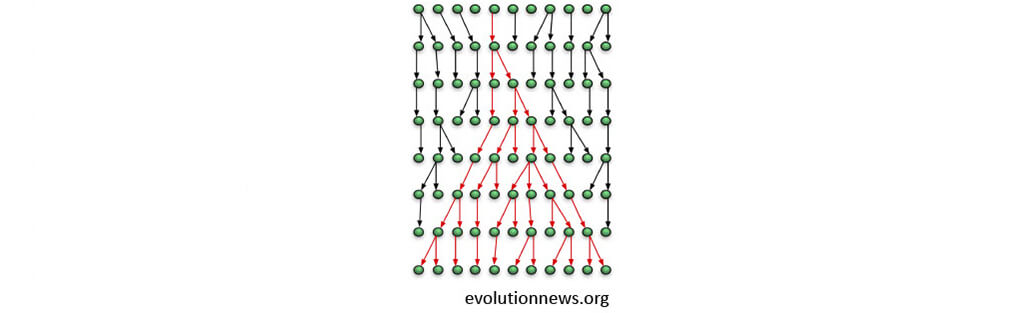
Many individuals do not produce any offspring
We can’t observe mutations in those individuals
Do all individuals have equal chance to produce offspring?
Did the mutation change the chance to produce offspring?
■Outline
- Basic concepts of genetics and evolution
- What is written in the DNA?
- How can we know what’s written in the DNA?
- How can we associate “genotype” with “phenotype”?
- Research on Thoroughbred horses
■What is written in the genome?
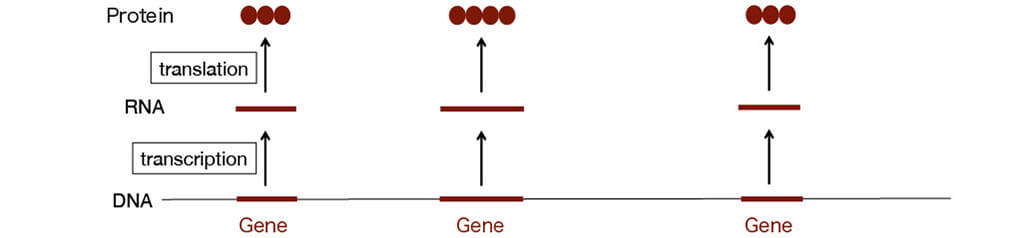
All cells contain the same set of genes
The amount and timing of RNA/proteins produced differ
(very complicated process)
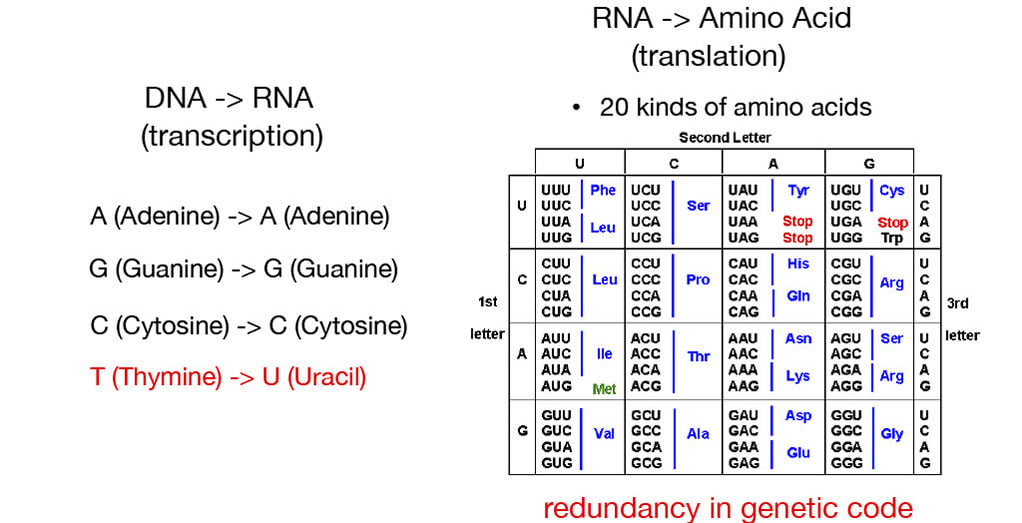
■Proteins as amino acid sequences
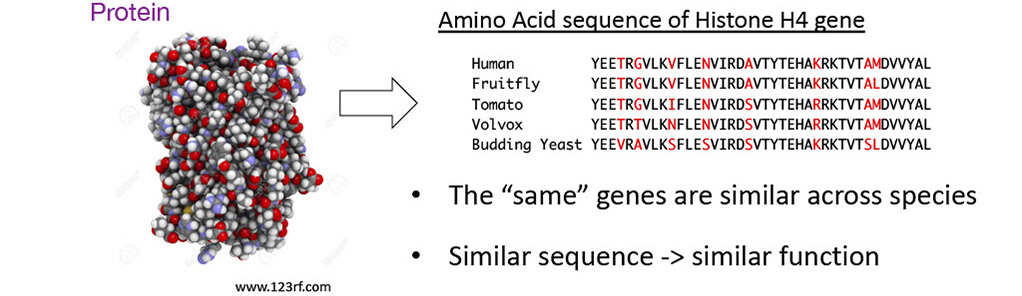
⇒Many genes share high similarity across distantly related species
■Gene structure

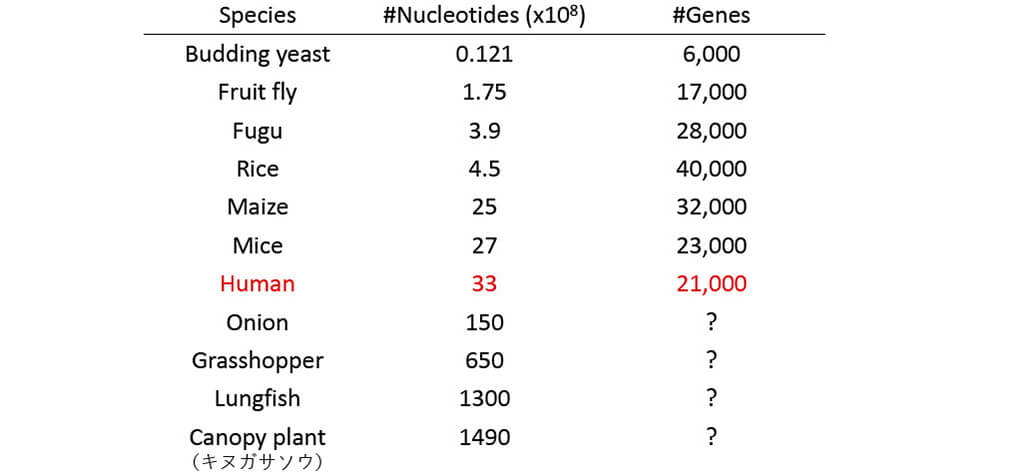
■Number of genes are not so different across species
■Evolution by Gene Duplication

- most genes were created by duplication of another existing gene
- mutation can create a new function while keeping the original function
■What is written in the genome?
- Human genome
protein-coding sequences: 1-2%
regulatory sequences: ~5-10%
Transposable Elements: >60%

- major reason for genome size variation
- virus-like parasites that amplify
- have their own “genes”
- often harmful, sometimes beneficial
■A lot of unnecessary information in the genome!?

“So much Junk DNA in our Genome” (Susumu Ohno, 1972)
We all acquire ~100 new mutations but most of us are fine (i.e. most mutations are harmless)
“Junk DNA” should be removed during evolution!?!?
- each “junk” is removed but lots of “junk” are being generated
- they are parasites that try to survive themselves
■Outline
- Basic concepts of genetics and evolution
- What is written in the DNA?
- How can we know what’s written in the DNA?
- How can we associate “genotype” with “phenotype”?
- Research on Thoroughbred horses
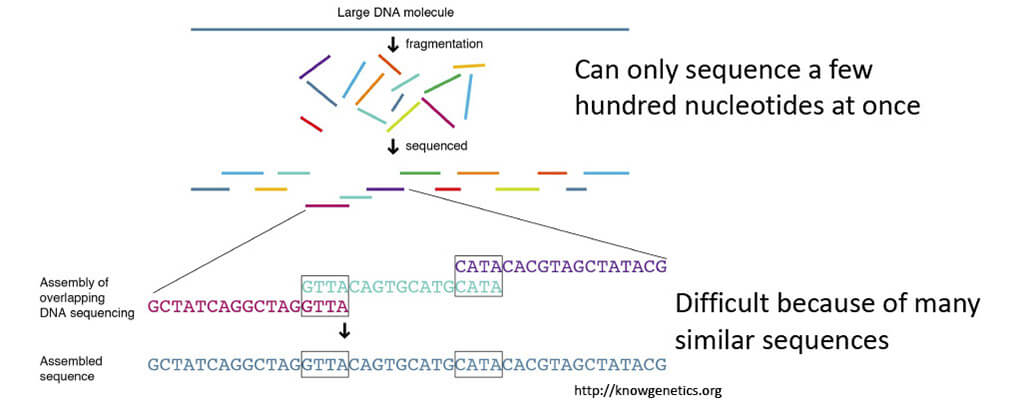
■How do we know the sequence of the genome?
- Genome sequencing and assembly
■How can we “find” the genes?

Extract RNA, sequence, and “map” them to the genome
Not so simple because of many similar genes and the presence of introns
Histone H4 gene
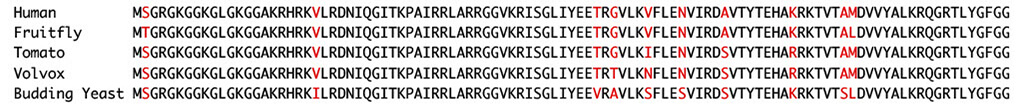
Search for similar sequences in the genome
Most genes in 1 species have similar genes in other species
Similar genes are likely to have similar functions
■How can we “find” the genes?
Predict genes based on their features

Nucleotide composition is statistically different
protein-coding sequence vs non-coding sequences,
introns GT-AG of intron vs GT-AG not associated with introns
■Outline
- Basic concepts of genetics and evolution
- What is written in the DNA?
- How can we know what’s written in the DNA?
- How can we associate “genotype” with “phenotype”?
- Research on Thoroughbred horses
■Domestication of Horses (~5500 yrs ago)

■Many diverse breeds established after domestication

- Thoroughbreds
- Originated in 18th Century (3 “founder” stallions)
- Horses that win races are selected to breed in many different countries
■Selective breeding of Thoroughbreds in Japan
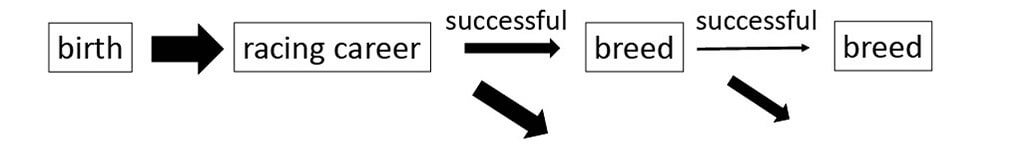
- ~7000 horses are born and registered at JRA (Japan Racing Association)
- Only 10-20 males per generation (>50% of females) are selected for breeding
⇒Thoroughbreds are much faster than other horse breeds due to selective breeding for 20-30 generations
==> but, genetic information has not been utilized yet
■Many differences between wild and cultivated Buckwheat

■Process of domestication

- Mutations occur that result in a desirable trait (might already exist in wild, or occur afterwards)
- Humans preferentially select and breed those individuals
- Can we identify the mutations selected by humans? (should speed up the selective breeding)
- Can we identify other useful mutations/genes that could be useful for breeding?
■Variation in the DNA of each individual
Single Nucleotide Polymorphism (SNP)

■Example of variation (SNP) associated with racing ability
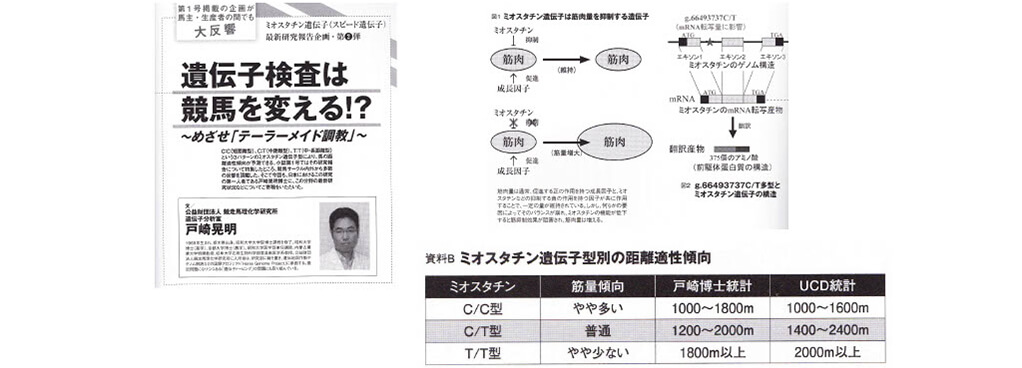
SNP in myostatin gene affects optimum racing distance
出典:サラBLOOD Vol. 1&2

■Using genetic information for Thoroughbred breeding
⇒Can evaluate the genetic potential of each horse
==> more informed choice of which horse to keep and which to discard
⇒Can identify the best breeding (male x female) combination
==> more informed choice of which male and female to mate

- ~400 Thoroughbred horses from JRA (Currently extended to ~1000 samples)
- Collect blood samples, extract DNA, identify SNPs
- Can we identify genetic variation associated with variation in traits?
- Can we identify genetic variation associated with variation in racing ability?
■Genome-wide Association Study (GWAS)

SNPs in neighboring region are “linked”
■GWAS works with coat color genes (proof of concept)
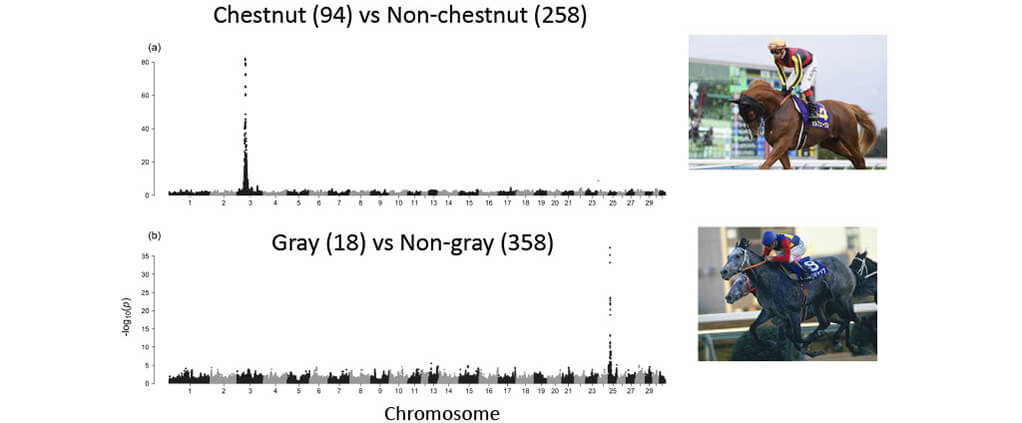
■Genomic signatures of selection
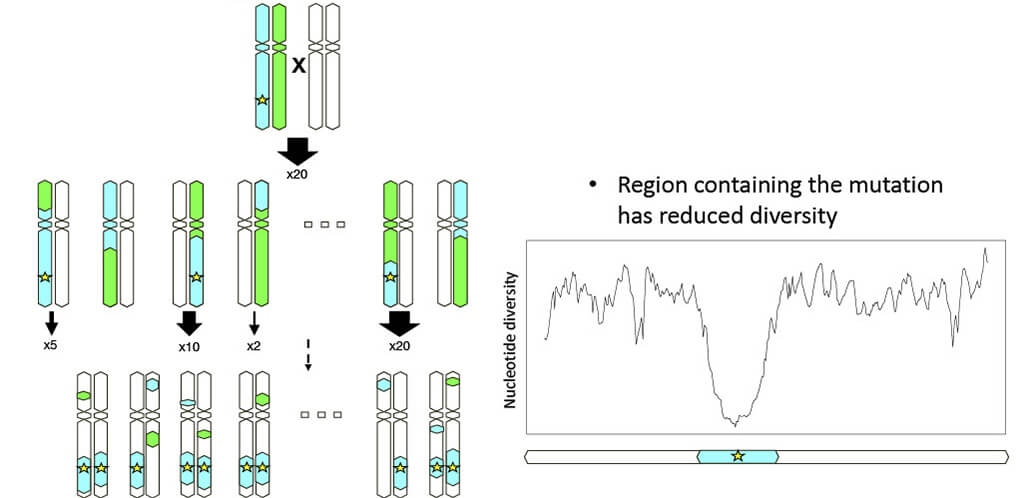
■Regions of reduced diversity regions in Thoroughbreds
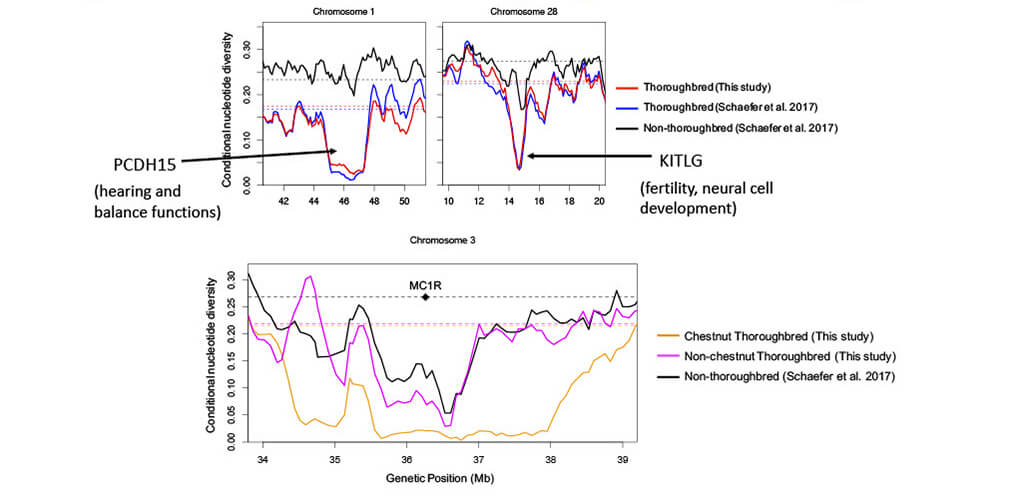
■Computational approaches to identify important regions
- Genome-wide association studies
very powerful when there is a targeted, simple trait - Genomic signature of artificial/natural selection
can use also for complex traits and where there is no specific trait- Difficult to pinpoint actual causative mutation/gene
- Can identify candidate regions/genes that can be tested experimentally
■Summary
- We can explain the process creating the diversity of life in a common
mathematical/computational framework - We can apply the same approaches to study many different species
(e.g. horses, buckwheat) - Computational/mathematical approaches are becoming
more important with the increase of data - More advanced mathematics are needed to go from “sequences”
to complex networks, shapes, 3D structures etc?
■Acknowledgements
- Thoroughbred project
Hideki Innan, Takahiro Sakamoto, Watal M Iwasaki (SOKENDAI), Fumio Sato (JRA Hidaka),
Teruaki Tozaki (Lab of Racing Chemistry), Takao Suda (Journalist), etc - Buckwheat project
Yasuo Yasui (Kyoto U), Chengyun Li (Yunnan Agri U), Takanori Ohsako (Kyoto Pref U),
Hideki Hirakawa (Kazusa DNA Res), etc
 ArithmerBlog
ArithmerBlog